School of Pharmaceutical Sciences, University of São Paulo; Ronin Institute · Funded by Grant #2019/26284-1 from the São Paulo Research Foundation (FAPESP).
This manuscript (permalink) was automatically generated from lubianat/quali_phd@f962072 on January 4, 2022.
The Human Cell Atlas (HCA) is an international effort to characterize every cell type of the human body.
HCA members are producing cell-level data from virtually all human tissue using techniques such as single-cell RNA sequencing, mass cytometry, and multiplexed in situ hybridizations.
The wealth knowledge stemming from the wealth of data needs management to maximize the societal benefit of HCA.
While semantic technologies have emerged as key players in the data interoperability ecosystem, there are still gaps to bridge.
Wikidata, a sister project of Wikipedia for structured data, is surfacing as a hub in the semantic web for multiple types of information and bridging experts and the people.
The connection of the Human Cell Atlas to Wikidata can quickly insert its products into the larger knowledge ecosystem, extending HCA’s reach.
This PhD project aims at studying Wikidata as a platform for representing cell types.
We review the literature on cell types, refining and formalizing concepts for cell type delimitation.
At the same time, integrate biomedical databases (e.g. PanglaoDB) into the Wikidata infrastructure and we enrich Wikidata with manually curated cell types.
To optimize the curation, we develop Wikidata Bib, a framework for literature management and organized note-taking system for dealing with the academic literature.
Finally, we are improving the interplay of Wikidata, the Cell Ontology and software used for single-cell RNA-seq data, inserting Wikidata de facto as a tool for the Human Cell Atlas community.
In this preface we present an overview of the chapters in this document.
The introduction contains an overview of the Human Cell Atlas project and the current state of classifying cells into types. Then, it introduces ontologies and knowledge graphs as tools for connecting what we know about cells.
The methodology section is an overview of the core methods used throughout the work. Of note, some chapters in the results section also display methods when they are intimately linked with the results.
It is worth noticing that the different results shown were not developed chronologically in the order shown. The different branches were developed in parallel, with overlapping periods of activity. They are organized into separate chapters and part of different publications.
The discussion on the concept of cell type is presented first, as it is instrumental for the later steps. It is followed by an account of how PanglaoDB, a database of cell markers, was integrated into Wikidata. Then, we present Wikidata Bib, a framework for an organized reading and curation of cell types. The framework, although used as a method throughout the PhD project, is also an intelectual product, thus presented in the results session. To end the results, we discuss how the work integrates with the Cell Ontology, the leading system for organizing cell types.
After the results, an account of other academic aspects of the project is presented as part of the qualification requirements. They present an overview of collaborations, participation in events and academic courses taken during the first part of the PhD project.
The Human Cell Atlas (HCA) project is, arguably, the biggest multinational biomedical project of the present time, running since 2017 to characterize every cell type in the human body [1].
The HCA consortium gathers people from all over the world to tackle different parts of the project to have a diverse and equitable account of the cell type diversity. [2]
It intends to “ultimately describe at least 10 billion cells, covering all tissues, organs, and systems” and, to achieve such bold goals, it commits to “to open membership, to the open and immediate data release with no restrictions, and to open-source code for all computational approaches”. [1]
Building a complete atlas of human cells comes with multiple challenges. The project includes the detection, in single cells, of RNA species (scRNA-Seq), chromatin accessibility (scATAC-Seq), and protein markers (primarily by CYTOF), as well as spatial information on cells with multiplexed in situ hybridization (such as MERFISH) and imaging mass cytometry [1,3]. The HCA is set to revolutionize the biomedical sciences by creating tools and standards for basic research, allowing better characterization of disease, and improving diagnostics and therapy. Its products (data, information, knowledge and wisdom) need to be FAIR: findable, accessible, interoperable and reusable. Data stewardship and management are growing as core demands of the scientific community, ranging from data management plans [4] to specialized data personnel [4].
The Human Cell Atlas has a dedicated team for organizing data: the Data Coordination Platform (DCP) [5] [3].
The DCP is responsible for tracing the plan for computational interoperability, from the data generators to the consumers.[3].
The Human Cell Atlas has its portal for data [6]), which composes the data repository landscape with other resources, like the Broad Institute Single Cell Portal [https://singlecell.broadinstitute.org/single_cell>) and the Chan-Zuckerberg Biohub Tabula Sapiens (https://tabula-sapiens-portal.ds.czbiohub.org/).
In addition to its core team, the HCA is poised to grow by community interaction.
As stated in its opening publication “As with the Human Genome Project, a robust plan will best emerge from wide-ranging scientific discussions and careful planning”.[1]
Thus, this project inserts itself among the wide-ranging scientific discussions to improve data - and knowledge - interoperability.
The highlight of “knowledge” in the last paragraph is meant to stress that raw data per se is not enough to turn the Atlas objectives into reality. There is a long way from raw datasets to commonly-agreed scientific knowledge. Currently, the gap between data and knowledge is attempted by writing and sharing scientific manuscripts, the de facto currency of exchange of claims about the natural world. A Human Cell Atlas Publication Committee reviews and selects publications, [7] presenting the papers as one of the significant outputs of the whole endeavour. As of december 2021, the list includes 96 different publications, which, arguably, expose only a fraction of the knowledge extractable from the underlying data.
The challenge that arises is one of managing a wealth of information and casting it into useful knowledge. Ideally, we would like to understand, remember, and use every statement produced by the HCA. As this goal is humanely impossible, we need to develop tools to make the knowledge interoperable with the aid of computers. At that point, the challenges of the HCA enter in resonance with the challenges of biocuration, and with the goals of this project.
One core step for knowledge management is the minting of identifiers for the concepts of interest. [8] The minting of identifiers, however, depend on clear, well defined entities. The sheer notion of “cell type” is undefined [1], a challenge for organizing both data and knowledge. Accordingly, this PhD project concerns itself also with the theoretical basis of defining a “cell type”. Thus, the next chapter will introduce the state-of-the-art of classifying cell into types, in a preparation for the chapter on knowledge modelling and the theoretical discussions of the results section.
Given that a core goal of the Human Cell Atlas is to advance knowledge about all human cell types, [1] the definition of “cell type” becomes essential. Although a number of views exist [9,10,11,12,13,14,15,16,17,18,19,20,21], there is no formal, commonly agreed upon defintion of cell type.
A 2017 article on the Human Cell Atlas mentions[12]: “Descriptors such as ‘cell type’ and ‘cell state’ can be difficult to define at the moment. An integrative, systematic effort by many teams of scientists working together and bringing different expertise to the problem could dramatically sharpen our terminology, and revolutionize the way we see our cells, tissues and organs. We invite you to join the effort.”
The article further highlights both the current gap in knowledge and the need for a community effort to work in finding definitions. The magnitude of the challenge justifies the position of the HCA to avoid attempting to propose a precies definition of ‘cell type’. [1]
One consequence of a lack of a definition is that there is no commonly agreed number of cell types, and not even on the order of magnitude. As of November 2021, the leading answers in the Google Search Engine for the question “How many different cell types are found in the human body?” all point to around 200 different types [22,23,24], an estimate that is agreed upon by Bionumbers, a database of useful biological numbers [25] [26]. A list of cell types in the adult human body on Wikipedia also amounts to around a couple hundred cell types [url?].
If we look at other sources, however, the estimates increase considerably. For example, the Cell Ontology has catalogued more than 2,311 cell types of interest for the Human Cell Atlas [27], increasing the estimate by at least one order of magnitude. Across the literature, we can find mentions of thousands to tens of thousands of cell types only in the brain. [28,29] Additionally, with an estimate of 37 trillion cells on average per human body [30] and an ever-increasing report of new cell types/clusters in single-cell transcriptomics [31], it is reasonable to assume the number of relevant cell types might be much larger. The Human Cell Atlas project itself does not commit to any estimates of numbers of cell types due to the sheer difficulty of estimating a number given current knowledge. (Aviv Regev; reply to a question in the HCA General Meeting conference [url?])
Even though there is no commonly agreed definition, different views on cell types are maturing, tailored to different research needs. One core line of thought to define “cell type” is based on the cell type as an evolutionary unit. That definition enables the drawing of parallels, from the evolution of other biological entities (such as genes, proteins, and species) to cell types’ evolution. Models of how multicellular life works greatly benefit from concepts as sister types (sharing a single ancestor), cell type homology (sharing a common evolutionary origin), and cell type convergence (executing similar functions without direct evolutionary links) [32,33]
Another school of thought is based on attractors: regions of dynamical stability in a feature space. [34,35] In this theory, “basins of attraction” direct cell phenotypes, providing points in, say, a gene expression space towards which different cells “move” their expression programs. This dynamic view sees each cell type corresponding to “a self-stabilizing regulatory program, which acts to maintain and restore the cell type-specific program of gene expression.” [36] It aligns itself with dynamic systems theory, and some authors go as far as to say that “Lacking the idea of attractors we have no clear idea of what a cell type is.” [37]
As much as different species concepts coexist [38], the quest to define cell types may take various forms. The challenge of representing cell types in evolution is conceptually different from representing cell types for identifying different entities on biomedical data. In that second direction, the groundwork of the Cell Ontology [39,40,41] is notable, providing identifiers that are reused across a range of databases. [42] Theoretical discussions on cell types is also the topic of specialized conferences, namely the International Workshop on Cells in Experimental Life Sciences conference series [43,44].
Even though many sources of knowledge contribute to our understanding of cell types [45], arguably single-cell transcriptomics is the workhorse for current efforts of the Human Cell Atlas. [45] Current scRNA-seq data analyses often rely on unsupervised clustering followed by labeling clusters. For the clustering, bioinformaticians select parameter sets to a target resolution, i.e., the level of detail used to detect cell identities. [46] [1] When the clustering is finished, the groups of cells are annotated with class labels, representing the underlying biology in a language we can understand. [47]
Instead of assigning expression gates, as done for flow-cytometry, single-cell RNA-seq analysis pipelines start from de novo clustering of cells followed by cluster annotation. [46] While it is clear that clusters and cell types are different concepts [46], often cluster labels are treated as cell types. There are several ways to cluster cells to find groups of similarity, and arguably the current default is derived from the methodology proposed by PhenoGraph. [48] The protocol is to calculate the distances between cells in a reduced PCA space (with the number of dimensions chosen by the experimenters), followed by constructing a k-nearest-neighbours network. Each cell is a node connected by k (another parameter) edges to other cells. Once the network is built, network modules (i.e. cell clusters) are commonly found using the Louvain algorithm, published in 2008 by researchers of the Université Catholique de Louvain in Belgium. [49] The cell clusters found by the PhenoGraph (or any other) algorithm are then labelled by domain experts, often based on genes called “cluster markers”, differentially expressed on each cluster. [46]
While it is possible to manually assign labels to clusters, automatic methods have been developed to aid in the task. [47] Mmarker-based automatic annotation bases itself on crossing clusters markers in the analyzed dataset with previous knowledge from databases like PanglaoDB [50] and CellMarker [51] [47]. Reference-based automatic cell annotation, on the other hand, relies on expert-annotated reference datasets to transfer labels to the dataset under study. [47] Other methods bypass the clustering step and focus on labelling individual cells, which avoids lumping different cells together. [47] Clarke et al.’s recent review and tutorial [47] provides an extensive account of current techniques for clustering and annotating cells.
Of note, even though a range of methods is available, most techniques and publications do not use standard identifiers for cell types. That omission disregards the advantages of using standard identifiers, such as those provided by the Cell Ontology. [47] [8] [27] [52] [53] [54]. Reassuringly, projects that use stable identifiers for single-cell RNA-seq data are appearing [42], including python and R packages (e.g. Besca [55], OnClass [56] and ontoProc[57]), data management projects and reference datasets, (e.g. Tabula Muris [58] and Tabula Sapiens [59] Azimuth map [60] and HubMap’s ASCT+B Tables [61]) and annotation platforms (e.g. the Cell Annotation Platform [62] and CellTypist [63]).
The Cell Ontology is currently the biggest provider of such standard identifiers. As elegantly put by Meehan et al. [64] the Cell Ontology is a “manually constructed computer-readable resource that links cell types by different relationships”. It was first described in 2005 [39] and was oriented both at creating a species-neutral classification of cells and for researchers to “learn a considerable amount about that cell type and its relationships to other biological objects” [39]:
The collaborative project gradually evolved and changed its design and scope to fit new needs.
By 2011, a need for computable definitions led to an advance in the number and quality of immune cell types represented in CL. [65]
It also included the addition of species-specific cell types to handle better marker-based definitions, usually presented at the species level. [65]
Further developments over the years included both technical improvements and the addition of new cell types.
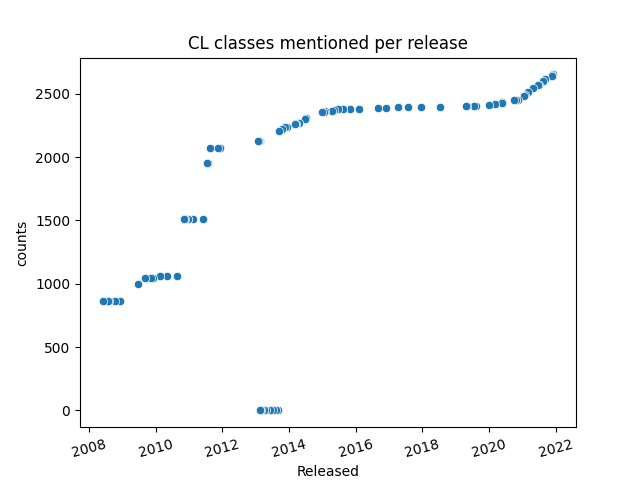
By the time of the last official CL publication, in 2016, it contained approximately 2,200 classes. [52]
Currently, the Cell Ontology is growing as a resource for the Human Cell Atlas and in providing identifiers for cell types [42] and presents over 2500 classes of cells (Figure 1)
The solution to the challenges of cell type classification, thus, pass via ontologies and other best practices of knowledge management. [42] Consequently, on the following chapter, we present tools for computer-based knowledge processing. We introduce the technical aspects of ontologies and knowledge graphs, present Wikidata, and discuss how such tools can can influence life-sciences research.
The classification of biological concepts is at the core of biology. At least since the Aristotelian endeavours to group classes of animals, a good part of the scientific work is to capture concepts into knowledge systems [66]. Linnaeus’ binomial system for naming species and Mendeleev’s periodic table are likely the two most famous classification systems but are part of a much larger ecosystem of structuring scientific knowledge.
On the 20th century, the development of the analytical philosophy of Russel and Wittgenstein and their search for formalizations [67] gradually layed the foundations for the the logic of scientific descriptions. Karl Popper and his “The Logic of Scientific Discovery”[68] was heavily influenced by analytical philosophy. Less known among life scientists, Tarski’s inquiries on what can be considered to be “true” [69] were also foundational for formal ontologies. In the end of the 20th century, the tradition of analytic philosophy contributed to the rise of applied ontology, which provided the theoretical basis for the computational ontlogies from the early 21st centhury. [70]
The whole movement for formalization of knowledge progressed on the computational end. At the late 20th century, the advent of computational ontologies and large-scale knowledge graphs were at the root of the functioning of the World Wide Web. This chapter will provide an overview of ontologies and knowledge graphs and their use in today’s biomedical sciences, alongside its future prospects.
An ontology, as used here, is a formal computational representation of reality, which tries to represent each concept (and their relations) as precisely as possible. [66]
Constructing an ontology is a process of selecting and defining terms of interest, selecting and defining relationships of interest and making statements about reality using terms and relationships.
The Gene Ontology is probably the most well known biomedical ontology; it describes (among other things) different classes of biological process related to each_other by “is_a” and “part_of relations. [71] [72].
The Gene Ontology is part of a much larger effort to formalize concepts across biology: the Open Biomedical and Biological Ontologies (OBO) Foundry. [73] Created in 2007, the OBO Foundry is a hub of biomedical ontologies that sets guidelines for designing and constructing high-quality ontologies. Under a common framework towards interoperability, the initial OBO Foundry united several independent ontologies (like the Cell Ontology, the Disease Ontology and the Protein Ontology). At the same time, the creation of the Relation Ontology (RO) provided a go-to point for relations in biology that different ontologies could reuse.
One of the OBO Principles for its ontologies is that they should be resolvable as a “syntactically valid OWL file using the RDF-XML syntax.” (http://www.obofoundry.org/principles/fp-002-format.html). The OWL Web Ontology Language was introduced as a standard by the W3C consortium in 2004. OWL is not a programming language, as it does not instruct computers to perform actions, but an ontology language, which allows computerizable descriptions of the world. Furthermore, it is an umbrella ontology language that includes several languages with varying levels of expressivity. Generally, more expressive languages can represent more complex ideas but make computations harder.
Regardless of ontology’s sublanguage, it must be resolvable to an RDF-XML file. RDF stands for Resource Description Framework, another W3C standard built around a graph-based data model [74]. Statements in RDF are triples consisting of 2 nodes (a subject and an object) and an edge (a predicate) connecting the nodes. All nodes and edges are represented in RDFs by International Resource Identifiers (IRIs), and there are many ways to lay out those IRIs on a text file to represent triples. One of those layouts is the RDF-XML syntax, inspired by the XML markup language. Arguably, other syntaxes (interchangeable with RDF-XML) are easier to read for a human. As an example of an RDF triple, here is how one would represent in the Turtle RDF Syntax, the notion that plasmacytoid dendritic cells are a type of dendritic cells:
http://purl.obolibrary.org/obo/CL_0000784 http://www.w3.org/2000/01/rdf-schema#subClassOf http://purl.obolibrary.org/obo/CL_0000451 .Where http://purl.obolibrary.org/obo/CL_0000784 and http://purl.obolibrary.org/obo/CL_0000451 are the unique IDs in the Cell Ontology for “plasmacytoid dendritic cells” and dendritic cells, respectively, and http://www.w3.org/2000/01/rdf-schema#subClassOf is the identifier for the “subclass of” relation as defined by the RDF schema.
A longer explanation of the details of OWL and RDF is outside the scope of this work. This brief introduction has a dual goal of introducing the architecture of formal representations and demonstrating the system’s complexity. There is a high energy barrier to acquiring the knowledge and the technical skills to engage in ontology building. That complexity might be one of the reasons why a tiny fraction of the biomedical communities represent data with ontologies, and an even smaller fraction engages with ontology building.
Even though the Semantic Web (which ontologies are a part of) spawned with promises of a revolution in the way knowledge is shared, it is still to be widely known outside semantic engineering. Two recent projects are playing a significant role in bringing the Semantic Web to a broader audience: the Google Knowledge Graph and Wikidata.
The Google Knowledge Graph introduced the Semantic Web de facto in the daily life of users of Google. [75/]. Its underlying structure is similar to the triples in an ontology, but it is less concerned with being logically coherent and does have strict semantics of a representation. In that way, Google Knowledge Graphs can feed on a variety of sources and not crash if there is some data modelling that, rigorously, could be inconsistent. Even though there is not a strict boundary between ontologies and knowledge graphs, one reasonable interpretation is that a knowledge graph may not be perfectly coherent, as long as it still can provide enough knowledge and reasoning for the approach of interest. While the lack of formal semantics limits reasoning and inference, the knowledge graphs are arguably easier to use, edit and understand, and so provide a user-friendly alternative for computable information with a lower entry barrier.
While the Google Knowledge Graph is widely used as a source of knowledge, it does not allow independent users to contribute information. On the other hand, Wikidata, the collaborative knowledge graph of the Wikimedia Foundation, allows users to contribute with classes and statements in the same spirit as Wikipedia and share its “epistemic virtues, like power, speed and availability. [76] Its power is derived from its large community of contributors, closely linked to the hugely successful Wikipedia. With a community of more than 20,000 active editors [77] and growing, it can cover a much wider number of concepts than any user individually. It is fast because one does not need to install any software or ask for permissions to update it: any user can do it via a web interface. That speed makes it easier for newcomers to join and contribute, in contrast to OBO Foundry ontologies, which require extensive training on semantics and knowledge of Git/GitHub for contributions. Finally, the information on Wikidata is available via a user interface, via a SPARQL query service and as large, full-size database dumps, providing full extent reusability. The Wikidata model has been so successful that Google decided to migrate its knowledge base, Freebase, fully into Wikidata.[78]
Wikidata uses the same framework (RDF) that powers ontologies, and its model represents statements about the world in triples containing a subject, a property and an object. [79] Its data model is serialized both in JSON and RDF. The data model contains 17 different data types, including, for example, “Item”, an entry on Wikidata that refers to “o a real-world object, concept, or event that is given an identifier in Wikidata” and “String”, a “sequence of freely chosen characters interpreted as text”. [80]. Knowledge is stored on Wikidata upon basic triples containing a subject (of type “Item”), a property and a value (which can be of any of the 17 types). As of November 2021, Wikidata contains more than 90 million data items [77] and more than 9000 properties that link them to values. As values often are other items, the database acquires a network format with labelled edges.
As seen in the example in 2, each of the items in the database contains an item identifier (Q followed by numbers). They also contain a label, a description, and a list of aliases, which can be recorded in more than 200 hundred languages, making it a multilingual project. [81] Each item is decorated with statements comprising property-value pairs. These pairs can be further specified via qualifiers and references, which treats the full triple as the subject, adding metadata to it (a process called reification [82/#reification]). Qualifiers provide ways to extend the information on the triple, while references provide provenance, enabling users to judge the validity of the claims in the database.
All the information is available on a user interface and programmatically. Advanced users can download dumps in JSON and RDF dumps and acess the data via the MediaWiki API and a SPARQL endpoint. [83] Several wrappers of such services are available in languages such as R [84] and python [85]. The data scheme can be seen in ?? model, where each item is connected to a statement node via a property in the “p:” namespace, from which references and qualifiers are accessible. To facilitate primary usage, the namespace “wdt:” connects items to values directly, simplifying, for example, the writing of SPARQL queries.
Information on Wikidata is released under a CC0 license, which enables full reuse of the data. [86] One of the major points of access and reuse of the information is the Wikidata Query Service [87], a core resource of the community which enables live querying in the SPARQL language. [88] A number of services make use of embedded queries from the Wikidata Query Service [87] to create interactive, live dashboards like Scholia [89] and the SARS-CoV-2 Query Book [90]
Wikidata is accessible in many ways and writable in many ways. It provides a user-friendly, point-and-click interface for modifying the database, providing low entry barriers for newcomers. It is also possible to semi-automatically reconcile spreadsheets to Wikidata items and use batch tools such as Open Refine [91] and Quickstatements [92], which enable batches on the magnitude of thousands of edits. For larger amounts of edits, it is possible to ask for bot permissions [93] and deploy systems that integrate big data sources. Bot edits are made via the Wikimedia API and are predominantly written via Python wrappers, such as Pywikibot [94] and the Wikidata Integrator. [95]
Due to its privileged position inside the linked data ecosystem and its ease of writing and query, Wikidata has been growing as a hub for interoperable data for the life sciences community. [96] [97] Even though Wikidata was created in 2013, the demand for a community-cured life sciences knowledge graph is apparent at least since 2008 [98] [99] The Wikidata-like project proposed was eventually discontinued, an example of the challenge of maintaining independent biomedical databases. [100] As Wikidata has a very large community, has stable funding and is at the core of modern technologies, like the Google Knowledge Graph [78] and Amazon’s Alexa, [101] it is virtually guaranteed that data in Wikidata will remain accessible for a long time, regardless of local funding schemes.
The Gene Wiki project [102] was likely the first large scale biomedical project to rely directly on the Wikipedia infrastructure for community curation.
It provided a direction connection between the generalist community of Wikipedia and domain experts.
The interplay of both communities is a topic of discussion and the opportunities and challenges were already discussed in NAR in 2012. [103]
Notably, Wikidata appeared chronologically after those efforts.
Notwithstanding, the Gene Wiki research group has embraced the Wikidata environment for community biocuration and data interoperability [104][105] [96] [106].
The information on Wikidata is still integrated to Wikipedias across multiple languages, often a a source of information in Wikipedia’s infoboxes.
Other projects outside the Gene Wiki initiative also started using Wikidata as a platform for knowledge integration. A list of several projects that use Wikidata as part of their service to their community is given in table ??. There is a movement exploring how Wikidata can be employed to advance Computational Biology and how it can be integrated to the current publication status quo. [107] In that direction, Wikidata is being developed as a platform for scholarly linked open data, mainly via the Scholia platform [108] [109],(https://scholia.toolforge.org/) which provides profiles of pre-templated SPARQL queries for entities like particular authors and articles (e.g. Scholia profile on Prof. Helder Nakaya available at https://scholia.toolforge.org/author/Q42614737).
| name | project website | citation |
|---|---|---|
| LOTUS | https://lotus.naturalproducts.net/ | [110] |
| GeneDB | https://www.genedb.org/ | [111] |
| Cellosaurus | https://web.expasy.org/cellosaurus/ | [112] |
| Complex Portal | https://www.ebi.ac.uk/complexportal/ | [113] |
| WikiPathways | https://www.wikipathways.org/ | [114] |
| Reactome | https://reactome.org/ | [115] |
| CIViC | http://www.civicdb.org | [116] |
| PubChem | https://pubchem.ncbi.nlm.nih.gov | [116] |
| Human Disease Ontology | https://www.ebi.ac.uk/ols/ontologies/doid | [116] |
During the COVID-19 pandemic, Wikidata has spawned as a hotspot for modelling information about the virus and the pandemic in real-time. [117] [wikidata:99196713?] The general scope of the database allowed representation in a shared system of molecular, epidemiologic and socio-economic aspects of the pandemic. [117][118] Information curated in Wikidata was immediately available, feeding live dashboards and other applications based on SPARQL queries. [119] [120] [121] Additionally, as the information presented on Wikidata is multilingual and collaboratively edited, it presented itself as a resource for constructing structured vocabularies in non-English languages. [122]
In addition to its value as a structured database, Wikidata is tightly connected to Wikipedia. The gene identifiers in the context of Gene Wiki [104] are now fed to Wikipedias across languages, benefitting users directly. Additionally, gene expression information from the Bgee database [123] was added to Wikidata and connected to Wikipedia, which led to a sizeable increase of access to the Bgee database. Currently, Wikipedia is one of the top 3 sources from which people access Bgee (personal communication with Tarcisio Farias), thus leading to direct recognition for integrated bases. More generally, the connections of Wikidata and Wikipedia make it unique in the power of flowing knowledge back to human-accessed interfaces. In the words of Matthias Samwald [124] and colleagues, “Wikidata could emerge as a community-backed and highly visible structured knowledge base of medical and biological information, bringing concepts and methodologies such as controlled taxonomies, Semantic Web / semantic technologies and ontologies into mainstream use.”
In conclusion, Wikidata’s unique position, robustness and guarantee of long term stability prompts the need for works exploring new ways of integrating it into current knowledge management systems. In light of the speed and breadth of the Human Cell Atlas and the challenges of knowledge representation on cells, this PhD works on addressing how Wikidata can play a role in organizing the discoveries about all human cell types.
Study and refine theories of classes of cells within the constraints of ontologies and knowledge bases
Catalog a comprehensive list of currently described cell types
Devise ways to connect life-sciences resources to Wikidata:
Provide proofs-of-concept of how Wikidata integration can benefit the advancement of the Human Cell Atlas Project
This project’s methodology resembles practical research-action practices [125]. The “action” facet is done by contributing to projects in the Human Cell Atlas and knowledge management context.
Research in 3 forms: - Philosophical investigation on knowledge representations of cell types, both in formal logic and in academic literature - Applied investigations of database integration and data quality in the context of Wikidata and biomedical ontologies - Data-driven biomedical research targeted at hypothesis generation and literature-based discovery using knowledge at the level of cell-type
All research branches are linked to the improvement of knowledge management in biomedical sciences, focusing on the Human Cell Atlas. The methods included the development and application of a framework for an organized reading of the scientific literature, providing contact with the different facets of biocuration and Human Cell Atlas-related research.
As much of the project is based directly on published research, we developed a reading framework, described in detail in the results section. The framework is based on GitHub and includes Python scripts, a file organizing the reading list, and another documenting the reading history in RDF. Notes and additional information are saved in a GitHub repository, and the structured information powers a live website with analytics on the users recent readings. The source code for Wikidata Bib is available at https://github.com/lubianat/wikidata_bib/tree/template and notes on my readings can currently be accessed at https://lubianat.github.io/wikidata_bib/.
Additionally, the methodology included a discipline of reading that entails the daily task of reading 2 articles, one about “cell types” and another about “biocuration”. The articles are obtained by a mixed manual and automatic approach, including a la carte selection of articles to read alongside Wikidata queries for Cell, Nature, Science and eLife papers about single-cell transcriptomics (query: https://w.wiki/4LHr) and papers on biocuration (query: https://w.wiki/4LHi).
For each article about cell types read, cell types previously absent on Wikidata are added via a combination of curation in a Google Spreadsheet and a custom Python script (https://github.com/lubianat/wikidata_markers/tree/master/curation_of_classes).
We have updated the Wikidata database with new entities, relations and triples. Creation of new entities was done in the Graphical User Interface (https://www.wikidata.org/wiki/Special:NewItem), via the batch Quickstatements tool (https://quickstatements.toolforge.org/#/) and the Wikidata Integrator python library (https://github.com/SuLab/WikidataIntegrator).
Properties, which link items to values need community approval. Under the scope of this PhD project, we have gotten the community approval for the properties : Cell Ontology ID (https://www.wikidata.org/wiki/Property:P7963) used to link cell types to their IDs in the Cell Ontology and has marker (https://www.wikidata.org/wiki/Property:P8872) used to link cell types to genes and proteins considered their markers.
The property acceptance cycle takes at least one week and is open for opinions by any Wikidata user. All the information regarding property proposals is available at https://www.wikidata.org/wiki/Wikidata:Property_proposal.
As part of the research-action process, I have joined the Cell Ontology working group. I participate in the monthly meetings and sporadic workshops, learning and contributing to the discussions. Additionally, I contribute to the ontology development, actively engaging in the Cell Ontology GitHub repository (https://github.com/obophenotype/cell-ontology) and contributing with new terms and assertions. I edit the ontology with the software for ontology editing Protégé v. 5.5.0 [126].
Status of cell type information on Wikidata was accessed via SPARQL queries combined with processing in python and is available at https://colab.research.google.com/drive/1GvQXOs51_U8icdGMtKXMeLOXKM8pXWet#scrollTo=szvBWI9zr_AA.
Counts of cell classes in the Cell Ontology were performed via regex matching on Cell Ontology releases following the code available at https://github.com/lubianat/cell_ontology_count.
As an initial step of this PhD project, we investigatet the definition of “cell type” for knowledge management on Wikidata. The definition of “cell type” is a topic of avid debate. [9,10,11,12,13,14,15,16,17,18,19,20]. Before we handled data in a large scale, we dedicated time for solidifying our theoretical basis of “cell type”.
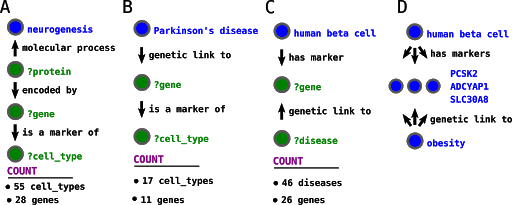
In a preprint derived from this PhD project [127], we proposed naming conventions for different cell types classes. Much of the literature uses the same names for single species (e.g., when dealing with a cell type as an evolutionary unit) or multiple species (e.g., in the Cell Ontology). Fe find it helpful to distil these different cases into different categories. Given the importance of the species’ concept in biological classification [128], we derive a species-centric view on naming classes of cell types. Using the notion of “taxonomic scope” as the taxonomic range in which instances of the class are expected to materialize, we propose 4 classes (Figure 4):
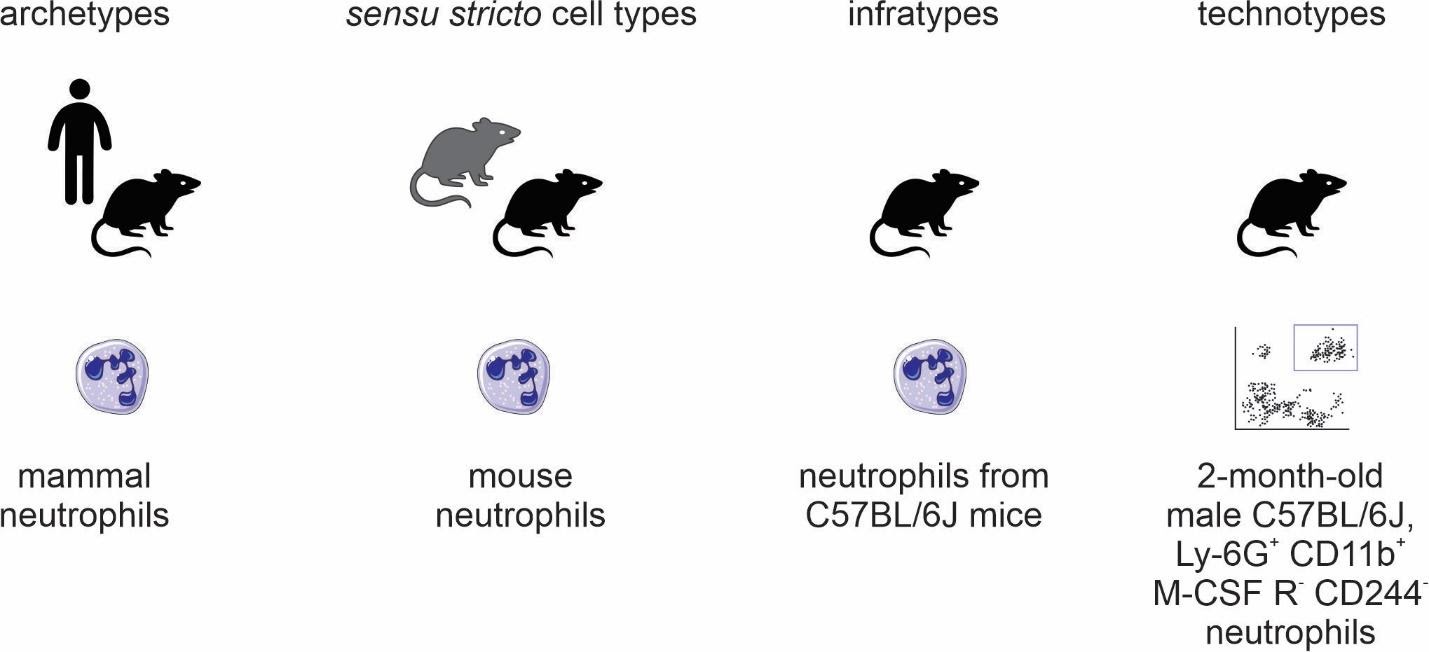
By specifying 4 different categories of cell types, we can hone in our organization.
While resources often do not make the taxonomic scope explicit, it might be inferred.
For example, the vast majority of biomedical articles present experiments with a single strain or a single species.
While two articles using different species might call some cells by the same name, they have important intrinsic differences.
Even if functionally equivalent, a mouse fibroblast can never become a human fibroblast, no matter the protocol.
While implicitly obvious, for computer-based, large scale knowledge management, it becomes necessary to state the obvious.
The division between archetypes and sensu stricto cell types is of particular importance for biocuration and data annotation. Usually, names and identifiers for genes and proteins are standardized for single species.[129] Thus, if we want to annotate marker genes, we would do better associating them to a species-specific cell type (a sensu stricto cell type) instead of the more vague association to a species-neutral type. Of note, current scRNAseq reference datasets and databases still use species-neutral cell type IDs(e.g. in the reference HuBMAP app; https://azimuth.hubmapconsortium.org/references/).
Our theoretical discussion on the notion of “cell type” extends the current state-of-the-art and introduces new ways to organize our knowledge about cells. The technotype and the infratype are currently pure theoretical constructs, and almost no resources deal with cell types at the level of strains or below. While we reason that this level of granularity would provide a more precise description of research projects, they are still far from being applicable in the present, and are present as tools for the future. The division of archetypes and sensu stricto cell types, on the other hand, was immediate value. As an example, it was instrumental for the integration of the Panglao database of cell markers to Wikidata, described in the following session of the results.
While theoretical discussion on the notion of “cell type” are important, a reasonable consensus is likely to take longer than the duration of this PhD project. Here we adopt a liberal view of cell type, defining, for our purposes, a cell type as any class of cells described by a domain expert with evidence of the reality of its instances. The requirement of evidence of existence in the material world is based on the principle of instantiation of ontological realism [130]. Barry Smith and Werner Ceuster state “A term should be included in a reference ontology only if there is experimental evidence that instances to which that term refers exist in reality. (‘Exists’ here should be understood in a tenseless sense in order to accommodate, for example, universals pertaining to extinct species as well as universals such as swarm or hurricane which are instantiated only intermittently.)” Following their advice, our minimum requirement for a cell type is public evidence for materializations of its instances.
By “type”, or “class”, we mean an abstract entity in a sense intended by the multilevel theory (MLT) of conceptual modelling [131] Figure 5 displays a simplified version of MLT adopted throughout this project. In this framework, real-world entities are materializations of individuals. Individuals are theoretical constructs that are (1) thought to exist or have existed, as per the principle of instantiation, and (2) refer to only one material entity at any point in time. For example, Wikidata’s entries for “Helder Nakaya (Q42614737)” and “Charles Darwin (Q1035)” are considered individuals by Multi-Level Theory. Other examples of individuals include “Albert Einstein’s brain (Q2464312)” and the “Christ the Redeemer statue (Q79961)”.
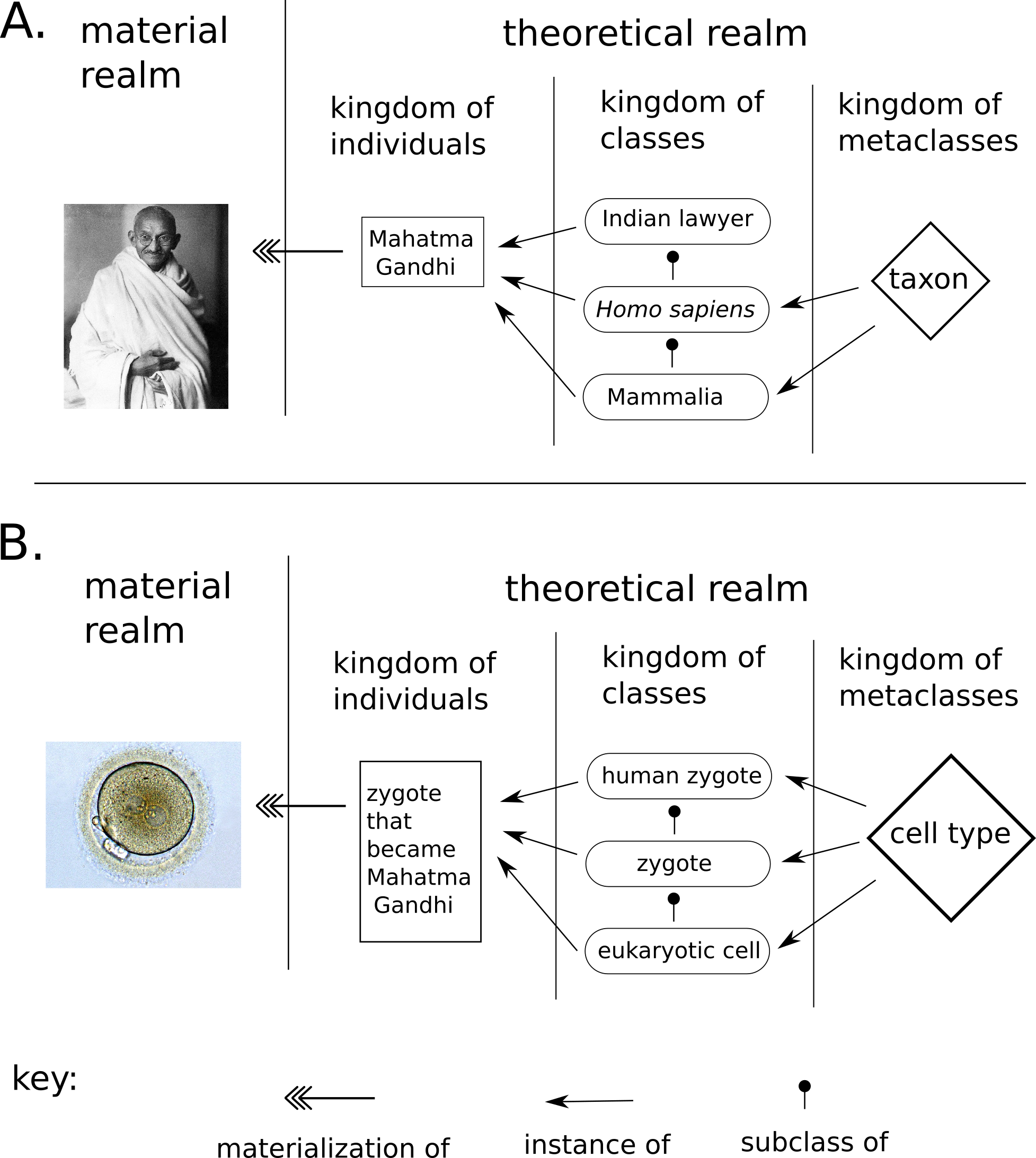
In MLT, individuals are instances of some classes. For example, both “Helder Nakaya (Q42614737)” and “Charles Darwin (Q1035)” could be represented as instances of the class “Homo sapiens (Q15978631)”. “Homo sapiens (Q15978631)” is only one of the classes that those individuals belong to. Another one is “animal (Q729)”. As all instances of “Homo sapiens (Q15978631)” are also instances of “animal (Q729)”, “Homo sapiens (Q15978631)” is a subclass of “animal (Q729)”. It is possible to continue the hierarchy of subclasses: “animal (Q729)” is a subclass of “organism (Q7239)”, and so on until the root case “entity (Q35120)”
Classes can also behave as individuals in some aspects. For example, both “Homo sapiens (Q15978631)” and “animal (Q729)” are instances of “taxon (Q16521)”. “Taxon (Q16521)”, thus, is a metaclass, or, more precisely, a 1st-order metaclass. Other examples of metaclasses are “species (Q7432)” and “phylum (Q38348)”. These, in turn, are instances of “taxonomic rank (Q427626)”, a 2nd-order metaclass.
In the Figure 5 B there is a proposal of this version of MLT for cell types. As individual cells are rarely named, for the sake of example, we can consider the “zygote of Mahatman Gandhi” as an individual in the theoretical system, an instance of the class “zygote (Q170145)”. “Zygote(Q170145)” is an instance of the metaclass “cell type (Q189118)” A more concrete example of from RNA-sequencing datasets, where there are barcodes for each cell in a particular sample. Each barcode labels an individual. Thus, labelling single-cells is a process of identification, where each individual is connected to a class of interest.
We avoid the dissection of the differences between persistent classes of cells (often called “cell types”) or the transient, fugacious classes of cells (often called “cell states”) (see “Definition of cell identity” section in [132] for an example). We also consider only the cell as it was observed in an experiment, not necessarily the future conditions of any cell (i.e., the “cell fate”). [35]
Even though such a distinction is an important topic for theoretical research, it is outside the initial scope of this work.
Another consequence of is that “subtype” becomes redundant with the idea of cell type and differ only stylistically. The notion of subtype, only will make sense in context of superclasses, but all in the same kingdom (i.e. the kingdom of classes).
We opted to frame our work around the term “cell type” due to its historical usage and familiarity for the life sciences community. The term “cell class” is also used in the literature and is a suitable synonym for our notion of cell type. Other related terms present semantic ambiguities, and were avoided whenever possible. For example, terms as “cell set,” “cell population,” and “cell cluster,” can reminisce of a specific, countable group of cells, frequently from the same experiment. The term “cell identity” has also been suggested for avoiding the cell type/cell state dilemma [46], but we avoid it to emphasize a nominalistic perspective (in the Popperian sense[133]). In doing so, we reinforce the intent to represent the cell types reported to exist instead of stating bluntly which cell types exist or are essential for human beings.
The employment of MLT and species-specific cell type are instrumental for the next chapters of this work. In the chapter about the PanglaoDB integration, we describe how we applied the theory to add marker information to Wikidata and cleaned up conceptual disarrays throughout the platform. Later, on the chapter about Wikidata Bib, we describe how we performed a large-scale curation of the biomedical literature for new cell types, using the theories discussed here as a starting principle.
Biomedical databases gather structured information for end users. They are present in different states of maintenance, and reconciling cell-oriented databases to Wikidata has the potential to increase interoperability, and multiply the value of previous biocuration efforts. PanglaoDB [134] [135] is a publically-available database that contains data and metadata on hundreds of single-cell RNA sequencing experiments. It provides extensive information on cell types, genes, tissues, and cell type markers, obtained via automatic and manual methods. It also displays a rich web user interface for easy data acquisition, including database dumps for bulk downloads.
As of 8 December June 2021, the article describing PanglaoDB had been cited 230 times. Despite its use by the community, the database is on a 3-star category for Linked Open Data [136] as it does not use the open semantic standards from W3C (RDF and SPARQL) needed for a 4-star rank, neither the links to external data via standard identifiers that make datasets 5-star. Improving the data format is a valuable step in making biological knowledge FAIR (Findable, Accessible, Interoperable, and Reusable). Thus, we provide a case study of making PandlaoDB available in a 5-star Linked Open Data Format on Wikidata.
As of August 2020, Wikidata had 264 items categorized as a “cell type”, considerably less than the Cell Ontology, which counts over two thousand cell types [52,137]. Strikingly, there were also 23 items categorized as instances of “cell (Q7868)”. This classification is imprecise, as an instance of cell would be an individual named cell from a single named individual, an example of conceptual disarray that often occurs on Wikidata. [138]
Wikidata editors often mix 1st-order classes such as “cells” and “organs” with metaclasses like “cell types” and “organ types”. As mentioned in the chapter on the concept of cell type and Multi Level Theory, individuals, like the “Dolly sheep zygote” and the “brain of Albert Einstein” are instances of classes like “zygote” and “brain”, respectively. Classes, like “zygote” and “brain” are instances of metaclasses, like “cell type” or “organ type”.
We diligently fixed and improved the conceptual consistency of cell type entries on Wikidata. As of 8 December 2021, the Wikidata database contains 2834 instances of “cell type” (see current status at https://w.wiki/b2t) and 0 instances of “cell” (https://w.wiki/4XAg) highlighting the improvements in both quantity and quality. This increase stems from the PanglaoDB integration (around 430 new types) and the Wikidata Bib curation described later.
After obtaining approval from the database owners, we matched genes and cell types to Wikidata and performed Wikidata queries to demonstrate the value of the approach.
An overview of the process is shown in ??.

Classes corresponding to species-neutral classes were curated from Wikidata using Wikidata’s Graphic User Interface. A manually-curated dictionary matching terms in PanglaoDB to Wikidata identifiers was assembled and used for integration. Cell types that were not represented on Wikidata were added to the database via the graphical user interface (https://www.wikidata.org/wiki/Special:NewItem) and logged in the reference table.
Species-specific cell types for human and mouse cell types were created for every entry in the reference table and connected to the species-neutral concept via a “subclass of” property (e.g. every single “human neutrophil” is a also “neutrophil”). Our approach was analogous to the one taken by the CELDA ontology to create species-specific cell types. [139]
After receiving authorization by e-mail from the PanglaoDB developer, Oscar Franzen, the PanglaoDB markers dataset was downloaded manually from PanglaoDB’s website (https://panglaodb.se/markers/PanglaoDB_markers_27_Mar_2020.tsv.gz) for integration. It contains 15 columns and 8256 rows. Only the columns species, official gene symbol, and cell type were used for the reconciliation.
The reconciled dataset was uploaded to Wikidata via the WikidataIntegrator Python package [95], a wrapper for the Wikidata Application Programming Interface.
Besides the Wikidata Dumps, Wikidata provides a SPARQL endpoint with a Graphical User Interface (https://query.wikidata.org/). Updated data was immediately accessible via this endpoint, enabling integrative queries integrated with other database statements.
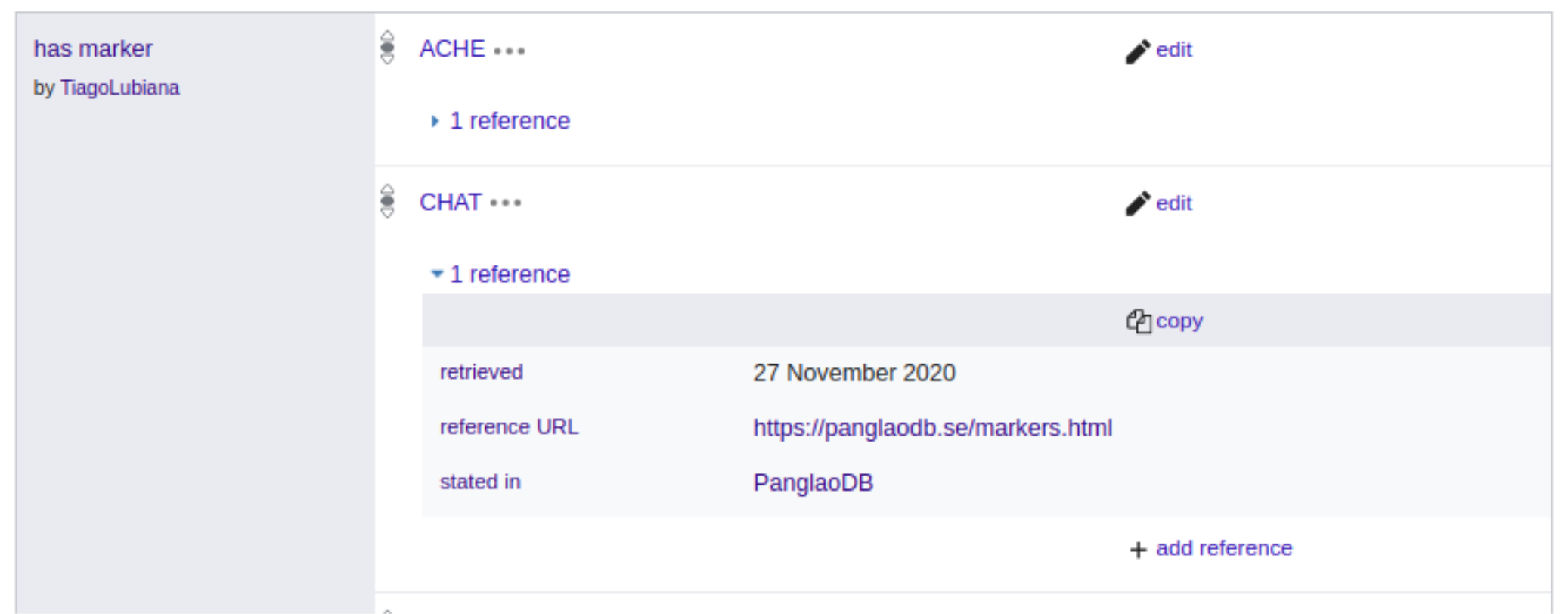
Adding marker information on Wikidata was not possible before this study and became possible after we proposed and got community approval of the property “has marker” (P8872). Figure 6 shows 2 of the current markers of “human colinergic neuron”(Q101405051), CHAT and ACHE, as they are seen on Wikidata. The PanglaoDB is referenced both via URL to the website (https://panglaodb.se/markers.html) and a pointer to the PanglaoDB item on Wikidata, Q99936939.
Now that we re-formatted the markers on PanglaoDB as Linked Open Data, we can make queries that were not possible before, including federated queries with other biological databases, such as Uniprot [140] and Wikipathways [141]. Due to previous similar reconciliation projects, Wikidata already contains information about genes, including their relations to Gene Ontology (GO) terms.
PanglaoDB’s integration to the Wikidata ecosystem allows us to ask various questions (figure 7).
As expected, the query below retrieved a series of neuron types, such as “human Purkinje neuron” and “human Cajal-retzius cell.” It also retrieved non-neural cell types such as the “human loop of Henle cell, a kidney cell type, and”human osteoclast. These seemingly unrelated cell types markedly express genes involved in neurogenesis, but that does not mean that they are involved with this process. The unexpected results reinforce that one needs to be careful when using curated pathways to analyze gene sets, as false positives abound.
The molecular process that gene products take part depends on the cell type. SPARQL allows us to seamlessly compare Gene Ontology processes with cell marker data, providing a sandbox to generate hypotheses and explore the biomedical knowledge landscape.
| geneLabel | cellTypeLabel |
|---|---|
| OMP | human purkinje neuron |
| OMP | human olfactory epithelial cell |
| OMP | human neuron |
| EPHB1 | human oligodendrocyte |
| EPHB1 | human osteoclast |
| PCSK9 | human delta cell |
| PCSK9 | human loop of Henle cell |
| CXCR4 | human b cell |
| CXCR4 | human T cell |
| CXCR4 | human NK cell |
Besides integration with Gene Ontology, Wikidata reconciliation connects the marker gene info on PanglaoDB with disease markers.
Disease genes are often compiled from Genomic Wide Association Studies, which look for sequence variation in the DNA. These studies are commonly blind to the cell types related to the pathophysiology of the disease. In the query below, we can see cell types marked by genes genetically associated with Parkinson’s disease. Even considering the false positives, the overview can aid domain experts in coming up with novel hypotheses.
| geneLabel | diseaseLabel | cellTypeLabel |
|---|---|---|
| BST1 | Parkinson’s disease | human b cell |
| BST1 | Parkinson’s disease | human neutrophil |
| RIT2 | Parkinson’s disease | human neuron |
| SH3GL2 | Parkinson’s disease | human alpha cell |
| SH3GL2 | Parkinson’s disease | human beta-cell |
In this part of the PhD project, we re-released the knowledge curated in PanglaoDB on Wikidata, connecting it to the semantic web. Each cell-type/marker statement was added to Wikidata with a pointer to PanglaoDB and a citation of the article, providing proper provenance. Based on the theoretical considerations on the concept of cell type, we added species-specific terms to Wikidata for cell types of Homo sapiens and Mus musculus described in the PanglaoDB database.
This work exemplifies the power of releasing Linked Open Data via Wikidata, and provides the biomedical community with the first semantically accessible, 5-star LOD dataset of cell markers, easily reachable from Wikidata’s SPARQL Query Service (https://query.wikidata.org/). Alongside other case studies of biomedical data integration to Wikidata (see [118], it contributes with tools and practices to serve as basis for contributors.) The work also paves the way for reconciling of other databases for cell-type markers, such as CellMarker [51], labome [142], CellFinder [137] and SHOGoiN/CELLPEDIA [143]) (if the owners give proper authorization). The approach we took here can be applied to any knowledge set of public interest, providing a low-cost and low-barrier platform for sharing biocurated knowledge in gold-standard format.
Reading scientific articles is an integral part of the routine of modern scientists. Although several literature-management software are available [144], the process of reading is mainly artisanal. There are no standard guidelines on how to probe the literature organize notes for biomedical researchers. Thus, while reading and studying is a core activity, there are few (if any) protocols for the efficient screening of scientific articles.
Other professional traditions have dealt with similar issues in the past. Notetaking is vital to keep track of financial balances and avoid costly problems in accounting. Double-entry bookkeeping was developed in the 13th century as a professional solution for notetaking in accounting where “every entry to an account requires a corresponding and opposite entry to a different account.” [145, =Double-entry_bookkeeping&oldid=1055066428] In software development, Test-Driven Development (TDD) is a popular methodology where tests for code snippets are written before the code itself, therefore ensuring that written software passes minimum quality standards. The similarities of Double-entry bookkeeping and TDD are diverse [146], but for our purpose, here suffices to see both as professionalized systems that promote better quality and accountability of works.
In the humanities, there is a well-established practice of annotations of readings. The annotation skills are part of standard academic training in the humanities [147][url?]. An influential work in presenting methods for academic reading in the humanities is Umberto Eco’s book “How to Write a Thesis” [148], which outlines not only how to annotate the literature that basis an academic thesis, but also why to do so. The book, written originally in 1977, is still influential today. Still, its theoretical scope (roughly the humanities) and its date preceding the digital era limits the extent to which it applies to the biomedical sciences.
Notably, the need for an organized reading system for biocuration studies stems from a difference in methodology. In humanities, the main (if not sole) research material is the written text, the books and articles from which research stems—[url?]. In the biomedical sciences, including a large part of bioinformatics, the object of study is the natural world, observed via experimentation. Thus, naturally, scientific training focuses on experimentation and data analysis’s theoretical and practical basis. With the boom of scientific articles, however, the scientific literature (and accompanying public datasets) already provide a strong material for sculpting scientific projects. Thus, developing a methodology for academic reading tailored to the digital environment is a need.
This chapter concerns itself with presenting Wikidata Bib, a framework for large scale reading of scientific articles. It is presented in three parts, each with a technical overview alongside the theoretical foundations. First, Wikidata Bib is presented as a reading system for managing references and notes using a GitHub repository and plain text notes. Then, we present how the system ensures accountability, allowing users to get personalized analytics on their reading patterns. Finally, we demonstrate how Wikidata Bib fits an active curation environment, connecting the framework with the larger goal of this project of curating information about cell types on Wikidata.
The reading framework of Wikidata bib is built upon a git repository integrated with GitHub, Python 3 scripts and SPARQL queries. It has a standard file structure, summarized as the following:
docs/
index.htmldownloads/
10.7554_ELIFE.52614.pdfnotes/
Q87830400.mdsrc/
get_pdf.pyhelper.pyread_paper.pyupdate_dashboard.pyindex.mdtoread.mdconfig.yamlpopwaddwadd_allwreadwlogThe docs/ directory contains the live dashboard from the readings, which will be discussed in the following sessions.
The downloads/ directory hosts the pdfs of the articles read with the system.
These are not committed to the repository and are only stored locally.
The notes/ directory contains markdown files, one for each article read.
The src/ directory contains the python code with the system’s mechanics.
They contain helper functions for the command line commands discussed below:
- wread which receives a Wikidata QID for an article and outputs (1) a notes document, (2) a pdf for the paper obtained from Unpaywall [149] and (3) an updated version of the dashboard HTML files in the docs/ directory.
- pop, which “pops” an article from toread.md and runs wread for it
- wadd, which takes an URL for a Wikidata SPARQL query and adds new QIDs to toread.md
- wadd_all, which parses config.yaml for recurrent SPARQL queries and runs wadd for each
- wlog, which adds, commits and pushes recent readings and dashboard updates to GitHub
All the structures described so far are commonly shared by any user of Wikidata Bib.
To personalize the use of the system, the user edits three plain text files.
toread.md hosts plain text QIDs of the articles that will be read.
These can be added either manually or via wadd.
While the pop command only sees QIDs, articles titles or other identifiers can temporarily be added to toread.md without breaking the system.
index.md hosts a numbered list of topics of interest.
This file plays the role of Umberto Eco’s work plan, with the topics of interest for the academic. [148]
These are used to tag articles for retrieval in a later step.
config.yaml contains shortcuts for different reading lists.
This is better explained by example.
In my to read.md file there are two reading lists, one following a # Cell types header and another following a # Biocuration header.
My config.yaml contains the following snippet:
lists:
# - shortcut: Title of header in toread.md
ct: Cell types
bioc: BiocurationThe config.yaml shortcuts are used as arguments by the pop command, where $ ./pop ct retrieves an article from the “Cell types” list, while $ ./pop bioc retrieves an article from the “Biocuration” list.
The Wikidata bib framework is coupled with a discipline of daily reading. The discipline is inspired by Robert Cecil Martin’s description of Test Driven Development in the book “Clean Code”, which includes not only a technical description but a school of thought of how software development might be approached. [150] Every day, I read one article of each list, using the notetaking station displayed in Figure [fig?]: notetaking. The constancy of reading allows steady coverage of the relevant literature. While the discipline has worked for this research project, it is not required to use the Wikidata Bib system.
The notetaking station of Wikidata Bib, opened in Virtual Studio Code, is depicted on Figure [fig?]: notetaking A.
The title and publication dates are displayed, and the reading process entails copying snippets from the text to the “Highlights” session.
Copying the highlights into plain text makes the sections of interest searchable via command line using grep (https://en.wikipedia.org/w/index.php?title=Grep&oldid=1039541979).
Comments can be added either in the comment section or inline, alongside the highlights, using --> Comment goes here to differentiate from highlights.
Also searchable by grep are the tags, copied and pasted from index.md in the ## Tags session or alongside the main article.
The discipline also includes, whenever possible, an improvement of the metadata about the article on Wikidata.
In [fig?]: notetaking B are shown the links included in the dashboard.
A link to a Scholia [108] profile allows identification of related articles from a series of pre-made SPARQL queries probing bibliography data on Wikidata.
While Scholia provides an overview of a given article, it does not allow direct curation of the metadata.
For that, two links are provided, one to Wikidata and one to Author Disambiguator [151].
By accessing the Wikidata page for the entity, one can add new triples, for example, curating authors and topics of the article, which are then used by Scholia and by Wikidata Bib’s dashboard.
Author Disambiguator is a wrapper of an Wikimedia API that facilitates disambiguating author names to unique identifiers on Wikidata, thus feeding the public knowledge graph of publication and authors.
Finally, a link to the article’s DOI or full-text URL is provided and serves as a fallback when the automatic download fails.
Of note, while the metadata curation has a technical benefit to Wikidata and the dashboard, it also plays a theoretical role.
By curating metadata on authors, the user of Wikidata Bib can better understand the people they read, and expand their metascientific perspective on their domain of interest.

The source code for Wikidata Bib is available at https://github.com/lubianat/wikidata_bib.
The Wikidata Bib system also enables the reader to get statistics on their readings.
Two simple databases are stored on the GitHub repository:
* read.ttl - An RDF document recording the dates in which each article was read.
* read.csv - An simple, human-readable index connecting QIDs with article titles.
The CSV file is only stored for accountability and as a quick way to glance at the titles read.
The .ttl file, on the other hand, is processed by the update_dashboard.py script to render 4 different HTML files under the docs/ folder:
- index.html
- last_day.html
- past_week.html
- past_month.html
All files are displayed in a GitHub pages.
In the case of this work, they are displayed at https://lubianat.github.io/wikidata_bib/.
To organize the code for rendering the dashboard, we created a python package, wbib, and deposited it in PyPi, making it available via pip. [152].
The package implements the logic for rendering complex Wikidata-based academic dashboards and is available in GitHub at https://github.com/lubianat/wbib.
It allows the user to build dashboards based on Wikidata records of information such as gender of authors, the region of author’s institutions, topics of articles and similar metascientific information.
The dashboard is composed of SPARQL queries written for the Wikidata Query Service [87]
It also allows users to feed an arbitrary list of articles and obtain a custom dashboard.
Wikidata Bib obtains the HTML dashboards after feeding wbib the lists of articles read in total (index.html) or in pre-determined time spans (last_day.html, past_week.html and past_month.html )
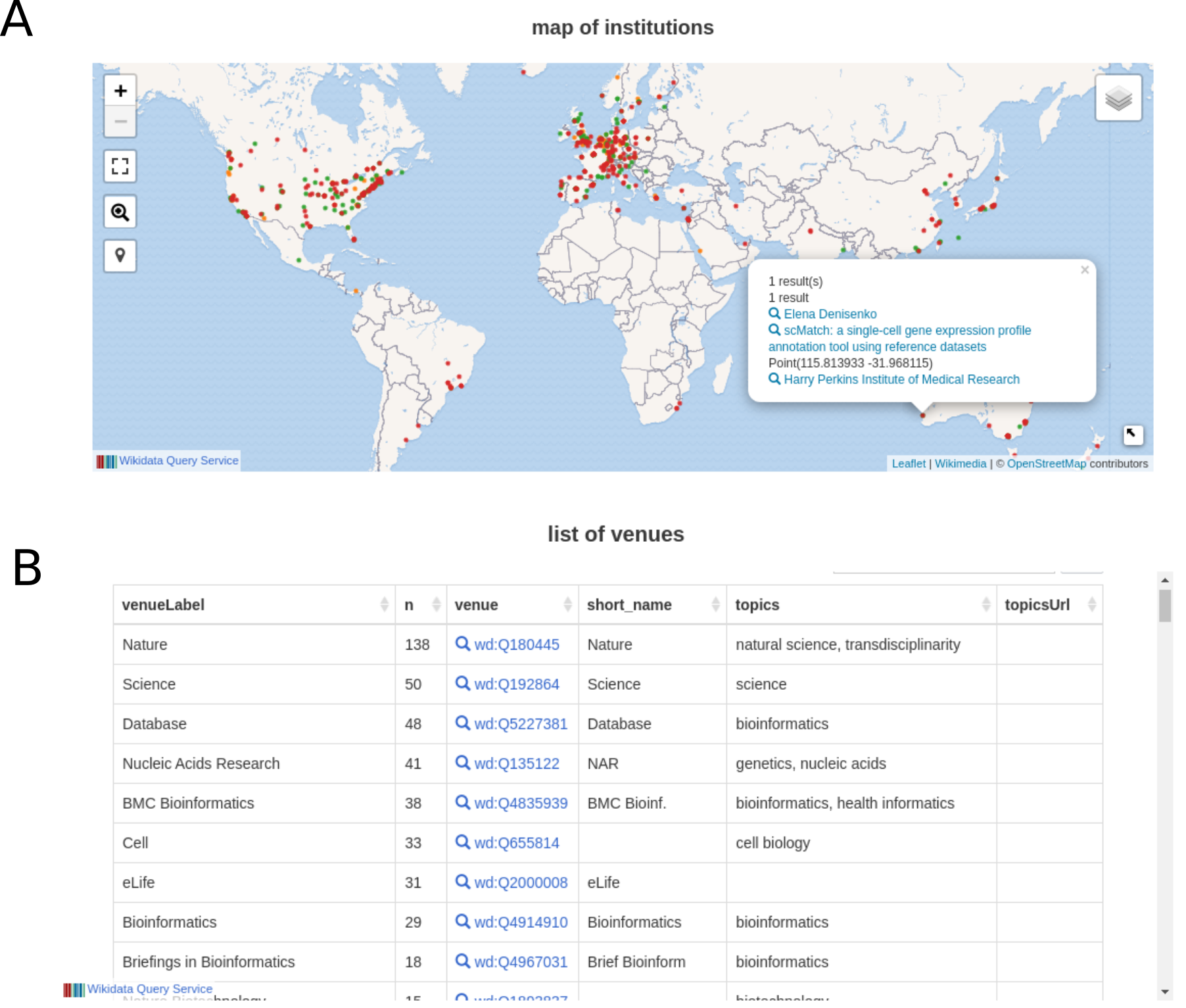
The dashboard includes not only a basic list of read articles, but also statistics on most read authors and most-read venues. It also displays an interactive map of the institutions of articles read, permitting a glance at geographic biases in activities. An example of queries is shown in 9. As the queries are rendered live, they evolve in quality with the growth of Wikidata. Finally, the clean 5-star-open data format enables users to adapt the queries to include different aspects of Wikidata. For example, table 4 showcases 10 articles that (1) I have read in the past year and (2) were authored by a speaker of the 1st Human Cell Atlas Latin America Single Cell RNA-seqData Analysis Workshop [153]. One practical application that the dashboard enables, thus, is to identify people in an event, institution or location that the user has read before, therefore catalyzing the possibility of collaborations. Anecdotally, this strategy was tested successfully at Biohackathon Europe 2021 [154], where I used the system both to identify possible collaborators and as a conversation starter.
| workLabel | authors |
|---|---|
| A promoter-level mammalian expression atlas | Jay W Shin |
| Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. | Muzlifah Haniffa |
| The Human Cell Atlas. | Musa Mhlanga, Jay W Shin, Muzlifah Haniffa, Menna R Clatworthy, Dana Pe’er |
| The Human Cell Atlas: Technical approaches and challenges. | Jay W Shin |
| Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. | Dana Pe’er |
| Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations | Sonya A MacParland |
| Single-cell reconstruction of the early maternal-fetal interface in humans | Muzlifah Haniffa |
| Distinct microbial and immune niches of the human colon | Rasa Elmentaite, Menna R Clatworthy |
| A cell atlas of human thymic development defines T cell repertoire formation | Muzlifah Haniffa, Menna R Clatworthy |
| Decoding human fetal liver haematopoiesis | Muzlifah Haniffa |
The Wikidata Bib system was devised originally to allow an overview of the fields of cell classification and biocuration. However, during the process, it was also repurposed for biocuration of new cell classes in Wikidata. By fast-tracking the reading of new articles, Wikidata Bib enables an efficient parsing of the literature and, thus, the identification of previously uncatalogued cell types.
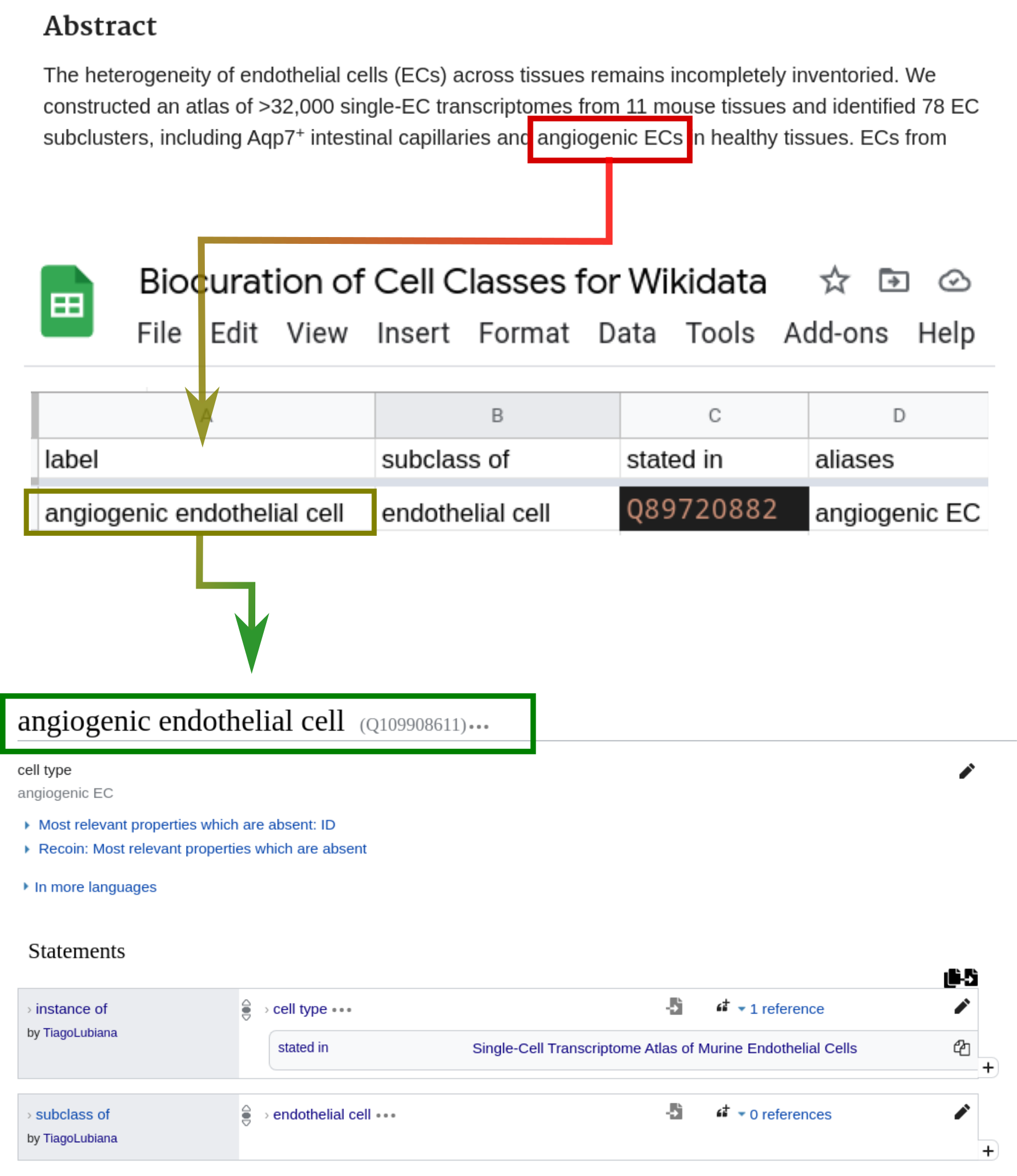
Articles read with Wikidata Bib were screened to mention cell types absent from Wikidata. As discussed in the chapter about the concept of cell type, we considered a “cell type” as any class of cells described by a domain expert with evidence of the reality of its instances. When a mention of such a class appears in an article, I first verify Wikidata for the existence of a related class. If it is absent from the platform, I enter a class name, alongside a superclass, and a QID in a Google Spreadsheet, as shown in Figure 10.
The information from the spreadsheet is pulled by a python script and processed locally with a series of dictionaries that match common terms to Wikidata IDs. In the example shown in Figure 10, the string “endothelial cell” was matched against a manually curated dictionary to the Wikidata entry Q11394395, the representation of that concept on Wikidata. After reconciling the data, the script uses the Wikidata Integrator python package [95] to insert the new entries on the Wikidata database. The code for integrating a Google Spreadsheet to Wikidata is available at https://github.com/lubianat/wikidata_cell_curation.
Wikidata contains 2940 subclasses of “cell (Q7868)” as of 8 December 2021.
From those, 550 cell classes are specific for humans, and 318 are specific for mice.
As a comparison, as of 8 December 2021, Wikidata has more cell classes than the Cell Ontology, which lists 2577 classes.
It is worth noticing that classes on the Cell Ontology are added after careful consideration by ontologists and domain experts and should be considered of higher quality than the ones on Wikidata.
From the 2940 cell classes on Wikidata, 2812 (95.6%) have been edited somehow by User:TiagoLubiana, and 1668 (56.7%) have been created by User:TiagoLubiana. Edits made to the cells were often connecting a dangling term, created automatically from an Wikipedia page to the cell subclass hierarchy, and included adding identifiers, images, markers, and other pieces of information. From the 1668 entities created, approximately 63 species-neutral cell types, 188 human and 188 mouse cell types were added based on PanglaoDB entries (total of 439). The remaining 1229 entries were created either via Wikidata’s web interface or via the curation workflow described in this chapter. These statistics are a simple demonstration of how the curation system efficiently contributes to the status of cell type information on Wikidata.
As mentioned by Aviv Regev in the Human Cell Atlas General Meeting 2021, after a shoutou to ontologies: “It’s everyone’s collective responsibility to participate in the annotation efforts, because that relies on domain expertise. To really tease apart things and give them names. Until we have names, people will have really a hard time working with things in biology.”[url?]” We hope that by developing simplified curation tools we will engage more domain experts into the curation efforts.
The contributions to cell types on Wikidata will be of most value if they are integrated to the current state-of-art of knowledge representation. Arguably, the Cell Ontology is the main source of cell type identifiers in the context of the Human Cell Atlas project.[42] Thus, data about cell types on Wikidata must be connected to the Cell Ontology.
To start the improvement in the interplay of both databases, we proposed and got the approval of a specific Wikidata identifier for the Cell Ontology, the “Cell Ontology ID” (https://www.wikidata.org/wiki/Property:P7963). IDs can be added to Wikidata entities and connected them to external databases enabling integrative SPARQL queries. Besides using the common Wikidata interface, one can crowd-curate identifiers via a 3rd-party service, Mix’N’Match, which provides a user-friendly framework for connecting identifier catalogues to Wikidata. [155/?p=114], as seen in Figure ??. Logically, we created a Mix’N’Match catalogue for harmonizing Cell Ontology IDs to Wikidata (https://mix-n-match.toolforge.org/#/catalog/4719), harnessing the community support for the task.

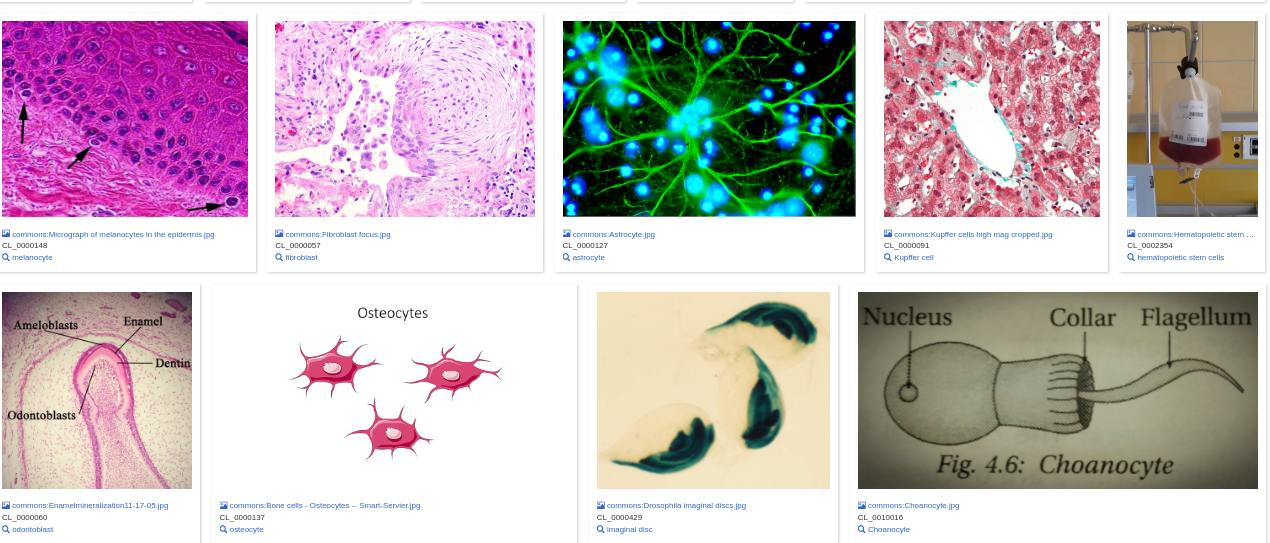
As of early December 2021, more than 700 Cell Ontology IDs have been manually matched to Wikidata. The integration already enables queries that harness the previously existing information on Wikidata for Cell Ontology-based applications. For example, one can query Wikidata items that have (1) a crossref to a CL ID (2) a picture in Wikimedia Commons (https://w.wiki/4F6e, Figure 12). The different possibilities of mutual benefit between the Cell Ontology and Wikidata will continue to be explored in the following years of this PhD project.
To sum up, this PhD research project aims at improving knowledge representation in the context of the Human Cell Atlas. It is composed of a mixture of theoretical studies on conceptual modelling, practical contributions to knowledge organization projects (mainly the Cell Ontology and Wikidata), explorations of the data to generate biomedical insights, and a technical framework for organized reading. By approaching the object of study from a new perspective, we hope to make sizeable contributions and promote discussion and fruitful conflation of approaches.
The next years of study will be devoted to improving the projects presented here into mature, useful objects. We hope to improve the interplay of Wikidata and Cell Ontology, developing frameworks to combine community- and expert-based curation of knowledge on cell types. Furthermore, we plan to integrate Wikidata to current single-cell RNA-sequencing pipelines by adapting R packages to use Wikidata (e.g. the ontology-based packages OnClass [56] and ontoProc[57]). Finally, we aim at moving the Wikidata Bib system to a well documented, user-friendly mature system, testing usability with other academics and distributing it as a durable open-source project.
During the initial course of this PhD work, we also completed the development and reporting of fcoex, an R package for investigating cellular phenotypes using co-expression networks. [156] The software was maintained to withstand new releases of dependencies and new R version and was published as a preprint on biorxiv. [doi:10.1101/2021.12.07.471603v1?]
Alongside the editing of cell-type information on Wikidata, I have joined different efforts to improve biological information on Wikidata. I have collaborated with the ComplexPortal curators as part of the Virtual Elixir BioHackathon 2020 (https://github.com/virtual-biohackathons/covid-19-bh20/wiki) and for the following year, to build a Wikidata Bot to integrate information on protein complexes to Wikidata. An overview of the Wikidata integration is in Figure 13, presented in an article published in Nucleic Acid Research (re-use of the image and legend possible under the CC-BY license of the article). [157]

I have also collaborated with the Cellosaurus database [158] to revive the CellosaurusBot [159], responsible for updating the metadata on more than 100,000 cell lines on Wikidata. The bot code, written in Python, was refactored entirely and runs semi-automatically after the Cellosaurus database was released. A write-up of the integration is in progress and is planned for release/submission in the first semester of 2022.
Finally, in collaboration with Olavo Amaral and Kleber Neves, from the Brazilian Reproducibility Initiative [160] I wrote a commentary on the value of publishing intermediate datasets as citable products. [161] The pieces discuss the value of small curations done both in systematic reviews and by experimentalists in the course of their research projects. Published curation tables can serve as a source for improving the ecosystem of open knowledge, not less by reconciliation to Wikidata (thereby bridging the commentary with this project)
During a part of this project, I have worked part-time as a consultant for the Wisecube company, based in Seattle, United States. [162] The job was approved by FAPESP and consisted mainly in writing SPARQL queries that probe Wikidata for answers to the questions posed by the BioASQ competition. [163] It also entails on-demand curation of biomedical topics on Wikidata based on requests by pharmaceutical companies as well as the development of dashboards targeted at providing insights to customers.
During the initial course of this PhD project, I have participated in several events:
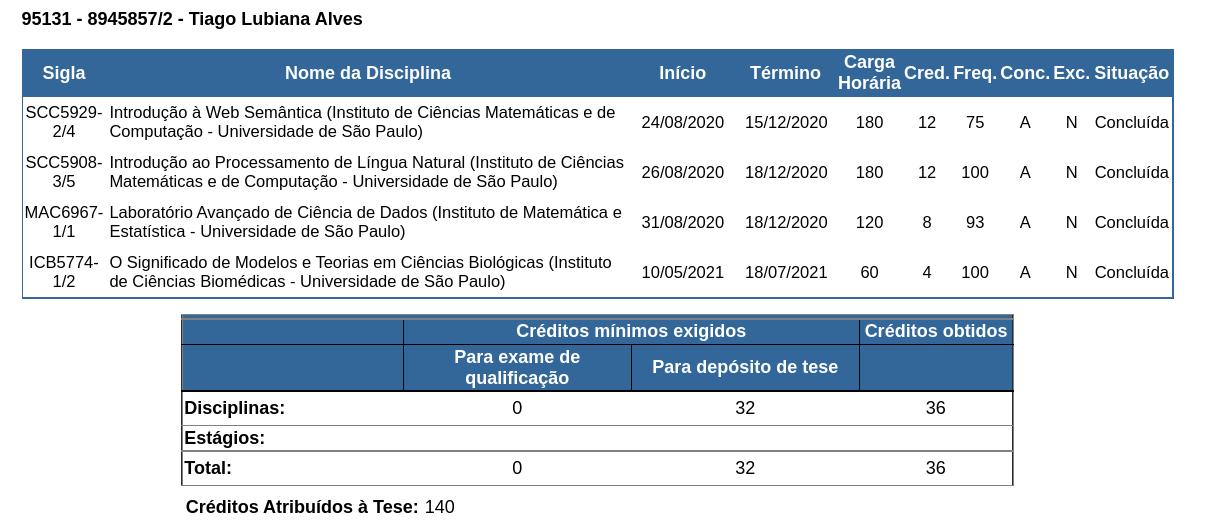
During the first year of the PhD program, I took four different classes, acquiring 36 academic credits. Figure 14 displays the disciplines taken, available only in Portuguese.